Prof. Douglas Thain at Notre Dame
Blog: Summer REU: Toward Elastic Scientific Applications
18 Oct 2010 - Douglas Thain
In recent months, we have been working on the problem of building elastic parallel applications that can adapt to the available resources at run-time. Much has been written about elastic internet services, but scientific applications have a ways to catch up.Traditional parallel applications are rigid : the user chooses how many nodes (or cores or CPUs) to use when the program starts. If more resources become available, or the application needs to grow, it is stuck. Even worse, if a node is lost due to a failure or a scheduling change, the program must be aborted. Rigid parallelism has been used for many years in dedicated clusters and supercomputers in the form of libraries such as MPI. It works fine for systems of tens or hundreds of nodes, but if you try to go bigger, it gets harder and harder to find a fully reliable system.
In contrast, an elastic parallel application can be modified at run-time to use greater or fewer resources as they become available, or if the size of the problem changes. Typically, an elastic application has one central coordinating node that tracks the progress of the program, and dispatches work units to other nodes. If new nodes are added to the system, the coordinator gives it some work to do. If a node fails or is removed, the coordinator makes a note of this, and sends the work to another node.
If you have an elastic application, then it becomes much easier to harness large scale computing systems such as clouds and grids. In fact, it becomes easier to harness any kind of computer, because you don't have to worry about them being reliable or even particularly fast. It's also useful in a traditional computing center, because you don't have to sit idle waiting for your ideal number of nodes to become free -- you can start work with whatever is available now.
The only problem is, most existing applications are rigidly parallel. Is it feasible to convert them into elastic applications?
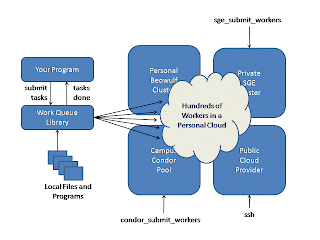
We hosted two REU students to address this question: Anthony Canino, from SUNY-Binghamtom, and Zachary Musgrave, from Clemson University. Each took an existing rigid application and converted it into an elastic parallel application using our Work Queue framework. Work Queue has a simple C API, and makes use of a universal Worker executable that can be submitted to multiple remote systems. A Work Queue application looks like this:
We hosted two REU students to address this question: Anthony Canino, from SUNY-Binghamtom, and Zachary Musgrave, from Clemson University. Each took an existing rigid application and converted it into an elastic parallel application using our Work Queue framework. Work Queue has a simple C API, and makes use of a universal Worker executable that can be submitted to multiple remote systems. A Work Queue application looks like this:
Anthony worked on the problem of
replica exchange
, which is a technique for running molecular simulations in parallel at different energy levels, in order to achieve a more rapid exploration of the energy landscape. Our friends in the Laboratory for Computational Life Sciences down the hall have developed a molecular dynamics engine known as
Protomol
, and then implemented replica exchange using MPI. Anthony put Protomol and Work Queue together to create an implementation of replica exchange that can run on an arbitrary number of processors, and demonstrated it running on Condor and SGE simultaneously hundreds of nodes. What's even better is that the computation kernel was simply the sequential version of Protomol, so we avoided all of the software engineering headaches that would come with changing the base software.
Zachary worked with the genome annotation tool Maker, which is used to do things like finding protein sequences within an existing genome. Maker was already parallelized using Perl-MPI, so this required Zach to do some reverse engineering to get at the basic algorithm. However, it became clear that the MPI aspect was simply doling out work units to each node, with the additional optimization of work stealing. Zach added a Perl interface to Work Queue, and converted Maker into an elastic application running on hundreds of nodes. We are currently integrating Maker into Biocompute, our local bioinformatics portal.
Zachary worked with the genome annotation tool Maker, which is used to do things like finding protein sequences within an existing genome. Maker was already parallelized using Perl-MPI, so this required Zach to do some reverse engineering to get at the basic algorithm. However, it became clear that the MPI aspect was simply doling out work units to each node, with the additional optimization of work stealing. Zach added a Perl interface to Work Queue, and converted Maker into an elastic application running on hundreds of nodes. We are currently integrating Maker into Biocompute, our local bioinformatics portal.
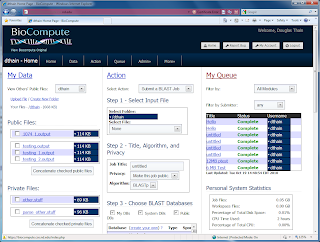
Speaking of
Biocompute
, Notre Dame student Brian Kachmarck did a nice job this summer of re-working the user interface to the web site. Not only is it faster and more visually appealing, it also does a better job of presenting the Data-Action-Queue concept described in our recent
paper about the system
.